Advancing Science-Based Approaches to Salmonella Control in Meat
Research and data-driven tools are being applied to reduce Salmonella risk in meat production and processing

Salmonella continues to represent one of the most persistent and multifaceted challenges in food safety, especially within meat production and processing systems. Despite decades of advancements in hygienic design, intervention technologies, and microbial monitoring, this pathogen remains the leading cause of bacterial foodborne illness globally. While prevalence in raw products has declined significantly, these reductions have not corresponded with proportional decreases in human illness.
Scientific evidence increasingly shows that the greatest risks stem from strain-specific factors such as concentration, virulence, and environmental persistence, rather than from presence alone. Modern molecular technologies such as whole genome sequencing (WGS), metagenomics, and deep serotyping now allow researchers and processors to examine Salmonella at a level of precision not possible a decade ago.1 These approaches provide insight into where and how contamination originates, how it behaves within production environments, and which control measures yield meaningful risk reduction.
This review summarizes recent peer-reviewed studies that advance our understanding of Salmonella ecology, detection, and control in meat systems and discusses how these findings inform future science-based approaches to food safety verification.
Applying Modern Technologies in Microbiology to Improve Food Safety
Traditional Serotyping
Identifies the dominant Salmonella serovar based on surface markers. Why it matters: useful, but it can miss low-level or slower-growing serovars.
Deep Serotyping
Uses sequencing to detect multiple Salmonella serovars in the same sample. Why it matters: finds emerging or high-risk serovars that routine methods overlook.
Metagenomics
Captures all DNA in a sample at once to reveal every microbe present. Why it matters: shows hidden reservoirs and mixed communities that traditional tests miss.
Whole Genome Sequencing (WGS)
Reveals all DNA of a Salmonella isolate, and comparison to others shows how strains are related. Why it matters: helps link what a plant finds to strains connected with illness.
Why This Research Matters Now
Traditional food safety systems have relied heavily on prevalence-based metrics, measuring whether Salmonella is detected rather than evaluating its potential risk. While this approach has been useful for establishing a baseline, we now know it provides limited information about public health outcomes because a simple positive or negative result does not reveal the concentration of the organism, the virulence of the strain, or its likelihood of contributing to human illness. Data from the Centers for Disease Control and Prevention (CDC) show that even as the proportion of Salmonella-positive samples has decreased, the overall incidence of human salmonellosis has remained relatively stable.2 This persistent gap highlights that controlling contamination alone is not enough; understanding which serovars and concentrations are most strongly associated with illness is essential.
To address this, the U.S. Department of Agriculture's Food Safety and Inspection Service (USDA-FSIS) has indicated a shift toward science-informed, outcome-based oversight that focuses on measurable reductions in illness rather than solely on prevalence.3 This approach aligns with the increasing availability of molecular and analytical tools that can link in-plant findings to national genomic surveillance databases, enabling more targeted and efficient interventions.
Recent Research
Microbiota Dynamics Across Pork Processing
Recent work examining microbial and pathogen behavior in pork processing plants has provided new detail on how microorganisms vary by line, shift, and location within a facility. One investigation using 16S rRNA sequencing showed that the microbial communities on meat and contact surfaces differed between fabrication lines and also changed across production days and times of day.4
Looking for quick answers on food safety topics?
Try Ask FSM, our new smart AI search tool.
Ask FSM →
Unique DNA sequence patterns appeared only during certain shifts. Although carcass lots were not tracked and could contribute to day-to-day microbiota differences, the repeatable shift-level patterns within each line indicate that in-plant factors such as work timing, sanitation, and line design were likely drivers of the differences observed.
Building on these observations, a review of literature by Asmus, Belk, and Noyes5 described how microbial communities shift from live animal through each processing step, supported by a schematic of the pork processing chain in Figure 1. Warmer, wet steps such as dehairing and polishing tended to support a wider range of environmental microorganisms, while colder areas like chilling and packaging were dominated by psychrotrophic taxa, meaning cold-tolerant groups such as Pseudomonas. Drains and equipment surfaces often carried microorganisms from one production period to the next, creating points where contamination could be reintroduced to carcasses.
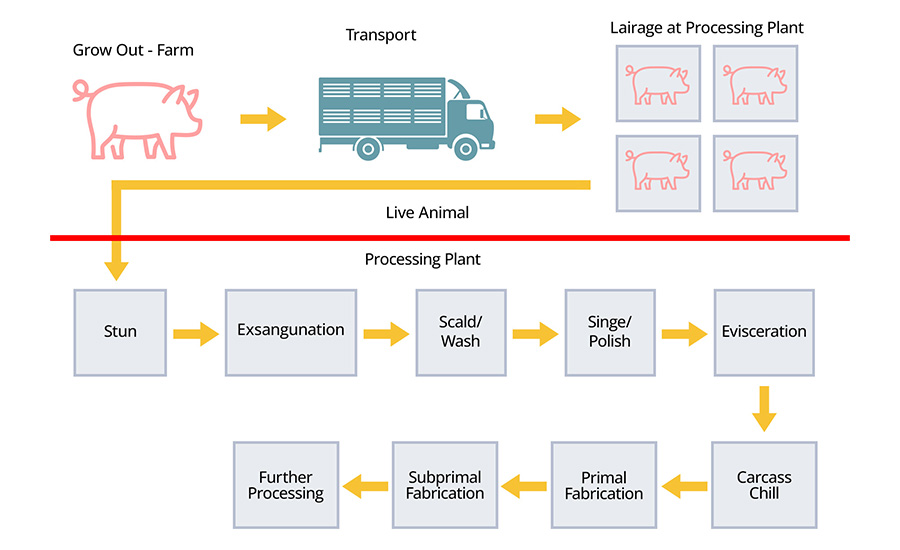
Another study examined how Salmonella moves and persists within a commercial pork plant by comparing isolates from two processing lines over multiple production days.6 Using whole genome sequencing (WGS), the researchers found that several strains were nearly identical genetically, differing by as few as zero to two single nucleotide polymorphisms (SNPs), which indicates they were essentially the same strain appearing repeatedly in different places and at different times. This pattern suggests that certain strains likely originated from a common upstream source and continued to reappear during fabrication, creating ongoing contamination that was usually present at low concentrations in individual samples and would be easy to miss without genomic tracking.
This pattern illustrates why pairing routine microbiological counts with strain-level genetic data is critical for understanding how Salmonella persists, rather than evaluating each sample as an isolated occurrence.
Serovar and Strain Differences
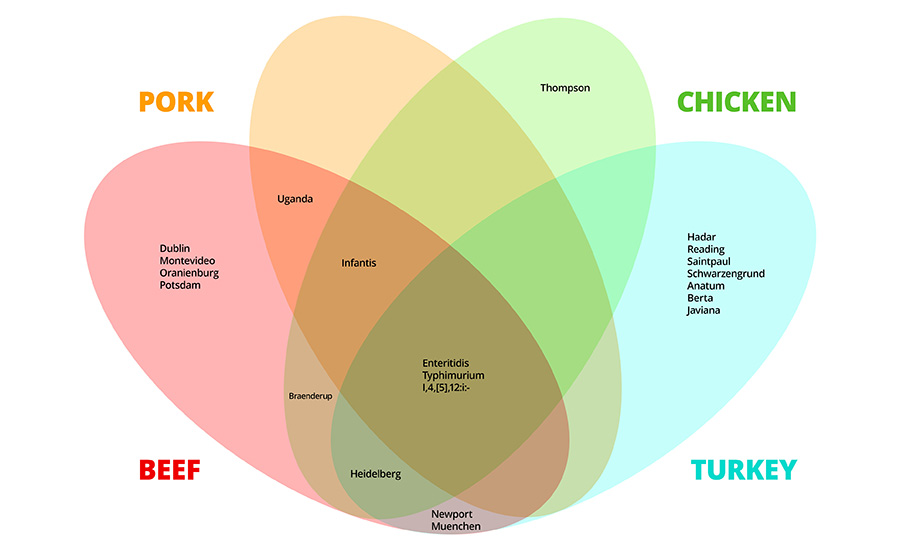
Advanced tools such as serovar–host linkage analysis, deep serotyping, and virulence screening methods like the Highly Pathogenic Salmonella (HPS) assay are possible only because of the extensive genomic datasets generated through the FSIS verification program, which has provided the foundation for developing practical, science-based tools for industry. Research continues to find that not all Salmonella strains pose equal risk. Serovars vary not only in virulence but also in host association and environmental persistence (Figure 2).7,8 Within processing environments, strain-level variation dictates how long an organism can persist and how effectively it withstands interventions.
For example, Salmonella serovar I 4,[5],12:i:- is consistently identified in national surveillance datasets as a high-concern strain because it is often multidrug-resistant and frequently linked to human illness, characteristics that can make it more difficult to manage within processing environments.7 While other serovars, such as London and Derby, are common, they display less persistence and lower virulence. The recognition of these differences is reshaping surveillance strategies; serotyping and sequencing data are now being used to rank serovars based on pathogenic potential. This information enables facilities to allocate resources toward mitigating the strains most likely to cause illness, rather than treating all positives as equal risk. Deep serotyping approaches are particularly effective at identifying minor or coexisting serovars within a single enrichment, providing earlier insight into emerging threats.
Quantitative assays that estimate concentration, rather than binary presence or absence, can better inform whether a process is moving toward or away from control. These findings have driven the development of tools that identify higher-risk strains more efficiently. Harhay and others7 have developed a method for screening Salmonella that are most virulent without having to first determine the serovar—the HPS assay. The method relies on detecting virulence genes that are most associated with the most pathogenic Salmonella, which is a faster and less expensive method compared with serotyping. Furthermore, as the most predominant serovars in different food animals can change over time, the HPS assay enables detection of the most important Salmonella without having to know the serovar.
Measurement at Low Levels
Accurate detection at low contamination levels remains a technical bottleneck for both regulators and industry laboratories. A recent U.S. Department of Agriculture (USDA)–Meat Animal Research Center study evaluated several culture-based and molecular approaches using poultry wing rinses. They found that although multiple methods could correctly identify samples at or above the 10 CFU/mL threshold, their consistency declined as concentrations approached the lower limits of detection. Some approaches tended to underestimate true levels near the threshold, leading to false negative results, while others performed more reliably once contamination exceeded 10 CFU/mL. These findings underscore the need for validated testing procedures with clearly defined decision rules when interpreting results close to detection limits.9
Together, these limitations at low contamination levels highlight the need to understand not only detection thresholds but also how different methods shape which serovars are identified. A separate study further demonstrated that enrichment bias and selective media composition significantly influence which serovars are detected.10 In some cases, minor serovars were completely masked by dominant competitors, leading to underestimation of overall diversity. Variability in detection of various serovars of Salmonella has implications for both prevalence reporting and root cause analysis, where incomplete detection may obscure true contamination sources.
Pre- and Postharvest Salmonella Control Gaps
Several ongoing challenges in preharvest and postharvest control of Salmonella in pork systems were reviewed, showing that certain biological reservoirs, including mesenteric lymph nodes, tonsils, and gastrointestinal contents, continue to complicate contamination control efforts.11
While preharvest strategies such as improved feed management, vaccination, and enhanced animal handling can reduce pathogen load, clear data on their effectiveness under commercial conditions are still limited. At harvest, interventions like hot water washes and peracetic acid rinses are well-documented tools, but their performance can vary depending on equipment design and carcass condition. Postharvest, persistent contamination in drains, conveyors, and trimming areas continue to present challenges. Progress will require validation studies conducted at true commercial scale so that the cumulative effect of multiple interventions can be measured, rather than evaluating each step in isolation.
Additional research illustrates how upstream biological factors influence contamination risk before animals reach the harvest floor. An additional layer of insight comes from recent work examining Salmonella in lymph nodes from cull sows and boars.12 Samples were collected from processing facilities located in the western and eastern regions of the U.S. during the winter, spring, and summer/fall months. The results showed that contamination patterns vary both by season and by lymph node type (Figure 3). Mesenteric lymph nodes were more frequently contaminated during spring and summer or early fall, while superficial inguinal lymph nodes had the highest contamination during winter in the eastern region of the U.S. Only the mesenteric nodes consistently contained Salmonella at levels high enough to quantify. These findings show how climate, geography, and tissue characteristics can influence Salmonella risk before animals even reach the harvest floor.
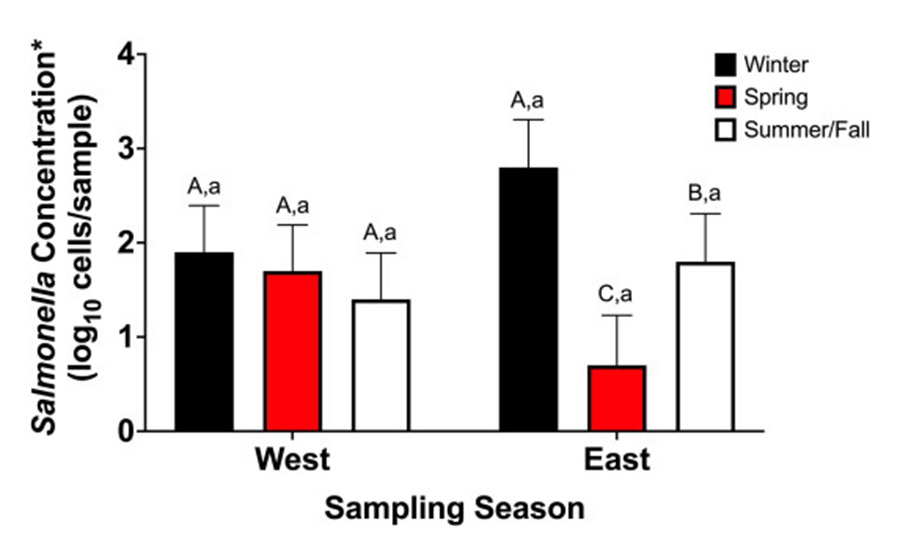
Notes: The season/region interaction was significant (P < 0.0001).a Superscripts that differ within a season indicate that regions are significantly different (P < 0.05).A,B Superscripts that differ within a region indicate that seasons are significantly different (P < 0.05).
Microbial Ecology in Small and Medium Processors
Small and medium-sized processors face distinct microbial challenges shaped by facility design, sanitation workflows, and resource constraints. Recent work examining microbial dynamics on belts, drains, and in the air of a commercial beef fabrication facility showed how bacteria persist, translocate, and recolonize surfaces throughout sanitation, underscoring that environmental reservoirs and sanitation-stage effects are critical considerations for plants of any size.13
Smaller operations often rely on multipurpose equipment and have shorter sanitation cycles, which can allow biofilms to develop on non-dedicated tools and surfaces. The study demonstrated that mixed microbial communities, including Listeria and Pseudomonas, can coexist with Salmonella in shared environmental niches, complicating elimination efforts. Routine environmental monitoring, using both culture and molecular assays, allowed early identification of persistent species before they were detected on product surfaces. This highlights the critical importance of accessible testing methods and basic microbial literacy among employees to recognize and interpret recurring contamination signals and take timely corrective action.
Practical Implications
The cumulative evidence from these studies provides a clear picture of where modernization of Salmonella control can have the greatest impact. Advancing control efforts may involve transitioning from point-in-time testing to more integrated systems that track contamination dynamics over time. Combining quantitative enumeration and strain-level data with biomapping approaches at both pre- and postharvest allows processors to understand not only whether contamination exists but also how and why it occurs. Data trends can inform when interventions lose efficacy, identify environmental niches that sustain recontamination, and supply the measured results necessary to refine process parameters based on observed performance. Ultimately, these tools support verification that aligns directly with public health outcomes rather than just compliance benchmarks.
Moving Forward with Science
The current body of research establishes a stronger scientific foundation for outcome-based Salmonella control. Integrating quantitative, serovar-specific, and genomic data into hazard analysis enables a more predictive and preventive approach to food safety. Collaborative initiatives among USDA research centers, FSIS, and academic partners are building data frameworks that connect genomic findings to practical interventions. For example, pilot projects are mapping virulence markers and antimicrobial resistance genes to specific processing conditions, allowing for targeted mitigation strategies that are measurable and replicable.
Recent validation studies also show that interventions can behave differently under commercial conditions than they do in controlled laboratory settings. McMinn and others14 demonstrated that the heat treatments used to destroy Salmonella, Shiga toxin-producing Escherichia coli (STEC), and Listeria in deli-style meats varied by product type and processing parameters, emphasizing the need for commercial-scale validation to ensure that interventions consistently deliver the intended level of lethality and process control.
As technologies like metagenomics, deep serotyping, and WGS become more accessible, processors can link facility-level data to public databases, creating a two-way flow of information that enhances surveillance and early warning. These same tools enable continuous feedback loops where intervention outcomes can be quantified in real time, strengthening both regulatory and operational accountability.
Ongoing research will remain essential to refining these systems and uncovering new solutions. Continued investment in science—spanning microbiology, data analytics, and process engineering—will help ensure that control strategies evolve in step with the pathogen itself. Further advancement will also depend on harmonizing preharvest and postharvest data streams and linking animal health management, processing interventions, and public health outcomes in a single continuum of risk assessment. By coupling scientific discovery with commercial validation and transparent data-sharing, the meat industry can make measurable progress toward reducing salmonellosis while reinforcing consumer confidence in product safety.
Turning Research Insights into Practical Action
- Persistent zones matter. Some equipment and facility areas consistently function as reservoirs for Salmonella and other pathogens. These locations should be incorporated into routine monitoring programs, with trends reviewed over time to understand persistence.
- Not all Salmonella are equal. Multiple serovars can appear in a single sample, and some carry higher public health risk. Methods that distinguish serovars allow control efforts to focus on strains most associated with survival or illness (Figure 4).
- Facility datasets may be underused assets. Environmental, product, and intervention data already being collected can reveal patterns when analyzed together by line, shift, equipment, or sanitation cycle. Even basic analytics can uncover meaningful risk drivers.
- Sampling is most informative at transition points. Processing steps where conditions change sharply often show the clearest shifts in microbial communities, providing early signals of contamination movement and intervention performance.
- Interventions function as systems. Validation under true production conditions clarifies how multiple control steps work together and where combined effects may weaken, guiding adjustments to sequencing or sanitation priorities.
- Higher-resolution typing strengthens decision-making. Deeper isolate characterization improves comparison with national pathogen datasets, supports root cause investigations, and helps separate true hazards from background flora.
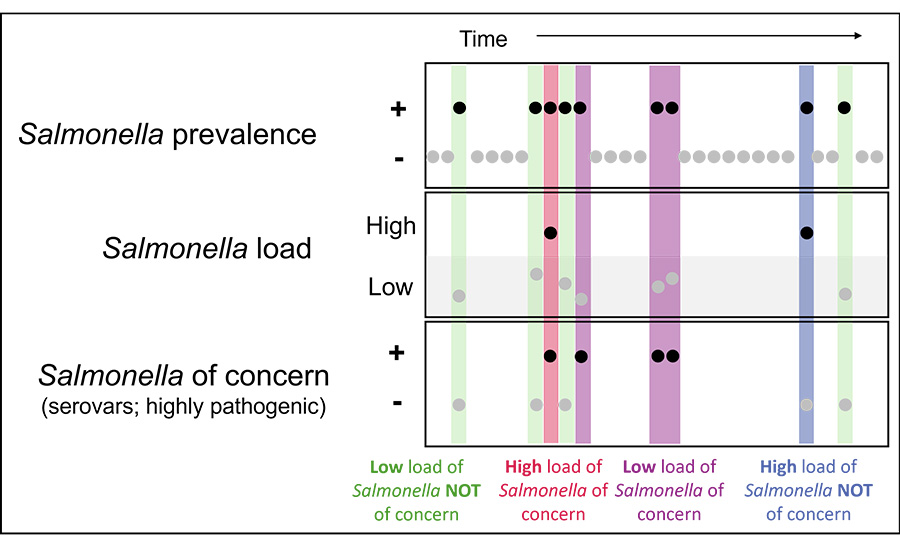
Note: This figure illustrates how three pieces of information work together when interpreting Salmonella results: whether Salmonella is detected, how much is present, and whether the strain is linked to illness. Different combinations of these factors signal different levels of concern and help guide how results should be interpreted for monitoring and control.
Acknowledgment
The author thanks Nikki Shariat, Ph.D., Associate Professor at the Poultry Diagnostic and Research Center in the Department of Population Health at the College of Veterinary Medicine at the University of Georgia, for her expertise and thoughtful review of this article.
References
- Shariat, N. "Serovar Differences Matter: Utility of Deep Serotyping in Broiler Production and Processing." Food Safety Magazine June/July 2024. https://www.food-safety.com/articles/9571-serovar-differences-matter-utility-of-deep-serotyping-in-broiler-production-and-processing.
- Centers for Disease Control and Prevention (CDC). "FoodNet 2023 Preliminary Data." https://www.cdc.gov/foodnet/reports.
- U.S. Department of Agriculture Food Safety and Inspection Service (USDA-FSIS). "Salmonella by the Numbers." Updated June 29, 2022. https://www.fsis.usda.gov/inspection/inspection-programs/inspection-poultry-products/reducing-salmonella-poultry/salmonella.
- Asmus, A.E., T.N. Gaire, K.M. Heimer, K.E. Belk, R.S. Singer, T.J. Johnson, and N.R. Noyes. "Fresh Pork Microbiota is Temporally Dynamic and Compositionally Diverse Across Meat, Contact Surfaces, and Processing Lines in a Pork Processing Facility."Applied and Environmental Microbiology 91, no. 4 (April 2025): e00044-25. https://journals.asm.org/doi/10.1128/aem.00044-25.
- Asmus, A., K.E. Belk, and N.R. Noyes. "Spatiotemporal Distribution of Microbiota During Fresh Pork Processing." Meat and Muscle Biology 9, no. 1 (2025): 20198. https://www.iastatedigitalpress.com/mmb/article/id/20198/.
- Asmus, A.E., K.M. Heimer, and K.W. Davis. "Temporality and Genetic Relatedness of Salmonella in a Pork Processing Facility." Journal of Food Protection 88, no. 5 (April 2025): 100500. https://www.sciencedirect.com/science/article/pii/S0362028X25000523.
- Harhay, D.M., K.D. Brader, T.S. Katz, G.P. Harhay, J.L. Bono, J.M. Bosilevac, and T.L. Wheeler. "A Novel Approach for Detecting Salmonella enterica Strains Frequently Attributed to Human Illness: Development and Validation of the Highly Pathogenic Salmonella (HPS) Multiplex PCR Assay." Frontiers in Microbiology 15 (January 2025): 1504621. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1504621/full.
- Katz, T.S., D.M. Harhay, J.W. Schmidt, and T.L. Wheeler. "Identifying a List of Salmonella Serotypes of Concern to Target for Reducing Risk of Salmonellosis." Frontiers in Microbiology 15 (February 2024): 1307563. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1307563/full.
- Schmidt, J.W., A. Carlson, J.M. Bosilevac, D. Harhay, T.M. Arthur, T. Brown, T.L. Wheeler, and J.L. Vipham. "Evaluation of Methods for Identifying Poultry Wing Rinses with Salmonella Concentrations Greater Than or Equal to 10 CFU/mL." Journal of Food Protection 87, no. 11 (November 2024): 100362. https://www.sciencedirect.com/science/article/pii/S0362028X24001467.
- Gorski, L., N.W. Shariat, A.K. Richards, A.T. Siceloff, A. Aviles Noriega, and D.M. Harhay. "Growth Assessment of Salmonella enterica Multi-Serovar Populations in Poultry Rinsates with Commonly Used Enrichment and Plating Media." Food Microbiology 119 (May 2024): 104431. https://www.sciencedirect.com/science/article/pii/S0740002023002186.
- Abrego, A., B. Wiseman, and S.E. Gragg. "Preharvest and Postharvest Salmonella Contamination in Pork: Current Knowledge and Gaps." Journal of Food Protection 88, no. 10 (September 2025): 100609. https://www.sciencedirect.com/science/article/pii/S0362028X25001619.
- Zhang, S., R.L. Brashears, M.M. Brashears, M.X. Sanchez, and S.E. Gragg. "Surveillance of Salmonella in Cull Boar, Sow, and Gilt Lymph Nodes and Tonsils from Six Cull Hog Processing Facilities in the United States. Journal of Food Protection 88, no. 12 (December 2025): 100642. https://www.sciencedirect.com/science/article/pii/S0362028X25001942.
- Yadav, B., Y. Fan, S. Hrycauk, T. McAllister, C. Narvaez-Bravo, T. Brown, and X. Yang. "Effects of Sanitation Practices on Microbial Dynamics in Meat Processing Environments." Journal of Food Protection 88, no. 12 (December 2025): 100647. https://www.sciencedirect.com/science/article/pii/S0362028X25001991?via%3Dihub.







.webp?t=1721343192)
