Monitoring All Salmonella Serovars in Poultry Production by Applying an Integrated Approach of PCR and HTS
Monitoring all Salmonella serovars is important to establish a complete epidemiological picture and prevent foodborne illness

Salmonella infection is one of the most commonly reported causes of foodborne illness, resulting in over 80 million cases of foodborne salmonellosis each year globally. Most illnesses are characterized by gastrointestinal symptoms such as diarrhea, fever, and abdominal cramps, with more severe illnesses leading to disability or death.1 While Salmonella contamination has been found in many types of foods, poultry and poultry products such as meat and eggs are well known to be the main sources of non-typhoidal Salmonella. Wild birds often carry Salmonella, which can be easily transferred among birds and via airborne movement of dust and fluff in poultry houses. Salmonella can be found in approximately 18 percent of birds, 22 percent of products, and over 29 percent of environmental samples, based on overall pooled prevalence of 157 studies from 15 countries.2 The Centers for Disease Control and Prevention (CDC) estimates that one in every 25 packages of chicken sold in grocery stores is contaminated with Salmonella.
Salmonella comprises over 2,600 serovars, which differ in their prevalence rate, transmission route, and pathogenic potential. Select serovars are frequently associated with human illness, while many are not. For example, a total of 131 serovars were identified from 157 studies carried out in 15 countries, with Salmonella Heidelberg, Kentucky, Enteritidis, and Typhimurium being among the most prevalent in poultry samples and those with the highest prevalence of anti-microbial resistance.2 While these serovars (particularly Salmonella Enteritidis and Typhimurium) are sources of numerous outbreaks, many other serovars have also been associated with outbreaks. Salmonella serovar prevalence also varies with location and over time. In the authors' recent study involving 192 poultry farms across Ontario, Canada, Salmonella Heidelberg, which showed high prevalence previously, has been drastically reduced.3 As a result, monitoring all serovars is important to establish a complete epidemiological picture to implement effective measures to prevent Salmonella transmission from poultry farms to consumers' tables.
Salmonella Proposed Framework Change
One of the primary vehicles for infection with Salmonella are eggs and egg-derived products contaminated with Salmonella Enteritidis. The bacterium is able to penetrate poultry reproductive organs, resulting in contamination of egg contents.4 As a result, regulatory agencies in many countries have established environmental hygiene monitoring programs for detecting Salmonella Enteritidis to identify potentially contaminated flocks for de-population and prevent contaminated eggs from reaching the market.
It is important to note, however, that other serovars can be involved in eggshell contamination. For example, 52 illnesses were reported across six states in 2015 due to eggs contaminated by Salmonella Oranienburg. Furthermore, 45 consumers from ten states were infected with Salmonella Braenderup in 2018, resulting in 11 hospitalizations and a recall of 207 million eggs throughout the U.S. Other serovars, such as Salmonella Typhimurium and Indiana, have been detected in eggshells collected from 41 farms in Australia.5 Many studies have also suggested that insects and animals can act as vectors for Salmonella introduction into poultry.6,7,8 As a result, in several jurisdictions eggs are required to be washed (sanitized) prior to release to market to mitigate the risk of eggshell contamination. In addition, infection of egg layers with certain serovars, such as Salmonella Gallinarum and Pullorum, can affect egg production and cause high mortality among flocks.9 Salmonella Typhimurium can also cause lesions that may lead to degeneration of oviduct and adversely affect food production.10
The current surveillance and regulatory programs are generally characterized by varied sampling practices and primarily rely on culturally confirmed results for regulatory enforcement. For example, in Canada, the Canadian Food Inspection Agency (CFIA) has a national Salmonella Enteritidis control program for the poultry industry; both CFIA and Salmonella Enteritidis insurance covered by the Canadian Egg Industry Reciprocal Alliance (CEIRA) require a confirmed culture result to make a claim for a flock depopulated due to a positive Salmonella Enteritidis test result.
In the U.S., the Department of Agriculture (USDA) and the Food and Drug Administration (FDA) have the National Poultry Improvement Plan (NPIP) with guidelines and specific measures to control the spread of Salmonella in poultry. This plan is voluntary, although most producers choose to participate for trading purposes. The NPIP also provides a cooperative industry, state, and federal program through which new diagnostic technologies can be effectively applied to the improvement of poultry and poultry products throughout the country. The NPIP was initiated in 1930s to help diminish the rampant spread of Salmonella Pullorum, which caused nearly 80 percent mortality in pullets. The program was later extended to include monitoring for other serovars, including Salmonella Enteritidis. The NPIP has approved or interim-approved 12 rapid detection methods, of which three focus on Salmonella Enteritidis, while the remaining methods focus on detection of Salmonella spp. and require culture isolation to determine a serovar.
In October 2022, USDA's Food Safety and Inspection Service (FSIS) released a proposed regulatory framework to reduce Salmonella contamination in poultry products. FSIS is evaluating whether establishments must consider Salmonella as a hazard and address Salmonella at receiving as part of their Hazard Analysis and Critical Control Point (HACCP) plans. In addition, the proposal would require that establishments monitor Salmonella levels or determine the serovars in incoming flocks. Under this proposed strategy, establishments would need to collaborate with their suppliers and contractors to ensure that they are implementing best practices for reducing Salmonella in breeding facilities, hatcheries, grow-out, and throughout transport. However, specific or additional rules, policies, methodologies, or protocols for implementing the strategy to achieve reduction goals are yet to be finalized.
Looking for quick answers on food safety topics?
Try Ask FSM, our new smart AI search tool.
Ask FSM →
Methods for Salmonella Detection and their Limitations
Standard methods for Salmonella detection involve pre-enrichment of a sample in a non-selective medium such as Buffered Peptone Water (BPW). The pre-enrichments are then subjected to polymerase chain reaction (PCR) screening or secondary enrichment in selective media, such as Rappaport Vasiliadis Soy broth (RVS) and Tetrathionate Brilliant Green broth (TBG), followed by plating onto selective agars such as Bismuth Sulphite (BS), Brilliant Green sulfapyridine (BGS) agar, and Xylose Lysine Deoxycholate (XLD) agar. Suspect colonies with morphological characteristics of Salmonella are then subjected to confirmation via biochemical reactions. A confirmed colony from a sample is then serotyped in a reference laboratory based on antibody-antigen interactions following the Kauffmann–White scheme,11 or whole genome sequencing (WGS) is used to determine its serovar.
The culture methods involve multiple steps that take several days to complete and require skilled technicians to perform and interpret. The current approach statistically favors the detection of the most abundant Salmonella serovars in a sample and misses less abundant Salmonella serovars, which can still be significant to public health. This discrepancy results in a partial and confusing epidemiological picture and compromises the effectiveness of preventive and intervention practices. Theoretically, testing more colonies from the culture plates would allow the detection of multiple serovars; however, this is practically prohibitive due to cost and labor increasing with testing an increased number of colonies.
Moreover, enrichment in different selective media, such as RVS and TBG broth, can lead to biased detection of certain Salmonella serovars.12,13 This issue becomes more profound when testing is required for identifying a specific serovar, such as Salmonella Enteritidis. The research is still limited to fully elucidate the impact that culture bias has on the probability of accurately identifying all Salmonella strains in a sample.14,15 The traditional culture-based serotyping method involves over 250 antisera to determine O (somatic) and H (flagella) surface antigens based on the Kauffmann–White scheme,11 and is performed mainly in reference laboratories due to demanding requirements of resources and technical skills. Polymerase Chain Reaction (PCR)-based methods can be used to detect a serovar without culture isolation, but it can detect only a limited number of pre-determined serovars at a time.16 The WGS approach still involves culture isolation and is mainly used in public health laboratories for pathogen subtyping.17,18,19
HTS for Detection of All Salmonella Serovars
The application of high-throughput sequencing (HTS) technologies, based on amplification of targeted identity regions, allows for the profiling of samples with diverse populations. PCR primers are used to amplify the region of interest from all target organisms in a sample, eliminating the need for isolation of pure cultures for identification purposes. Following HTS, robust data analysis tools are used to sort sequences and assign identity to known taxonomic groups, facilitating the detection of organisms of interest in a sample and also providing relative abundance information. The use of PCR and HTS can circumvent the need for further enrichment and isolation steps required by standard culture methods in Salmonella detection and serotyping, as they provide the ability to profile all Salmonella serovars present in a sample by sequencing an identity region contained in all Salmonella genomes.
Salmonella genomes contain Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) sequences that can be used to differentiate among serovars. A recently reported method called CRISPR-SeroSeq (serotyping by sequencing) employs HTS to profile Salmonella serovars in a sample by targeting a defined region containing the CRISPR loci in the Salmonella genome.20 The direct repeat regions are conserved and used as flanking sequences for PCR amplification. Publicly available Salmonella CRISPR sequences are used to construct a database to assign a Salmonella serovar identity to unknown sample sequences generated upon HTS of indexed PCR amplicons. The number of sequences assigned to particular Salmonella serovars enables both the detection and calculation of relative abundance of multiple Salmonella serovars in an individual sample, including both the predominant and less abundant serovars present in mixed populations. The CRISPR-SeroSeq method has been demonstrated to have a similar detection sensitivity and reproducibility as PCR, as validated using poultry environmental samples. It is also able to simultaneously detect common Salmonella serovars, with minority serovars being detected at abundances as low as 0.01 percent in a mixed Salmonella population.3
Complete Picture of Salmonella Serovars by HTS
The use of CRISPR-SeroSeq to profile Salmonella serovar populations offers a more complete picture and better understanding of shifts observed in Salmonella serovars detected at various stages of poultry production, as demonstrated in a recent study conducted by Siceloff and colleagues.21 The study revealed Salmonella serovar prevalence in broiler carcasses and raw chicken parts from 2016–2020 in the U.S. and identified regional differences in Salmonella serovars, with the Atlantic and Southeast regions more frequently isolating Salmonella Typhimurium compared to the South-Central, Midwest and Mountain, and West regions.
The data from surveillance at different stages of production also revealed that across regions, Salmonella Kentucky decreased in relative proportion within samples from carcass to raw chicken parts, while Salmonella Enteritidis had the opposite trend. In Georgia, Salmonella Kentucky isolation increased between 2016 and 2020 in not only carcass samples, but was also the most frequently isolated serovar in samples taken from breeding flocks at 15–19 and 40–45 weeks of age. Other serovars, such as Salmonella Enteritidis, Typhimurium, variant I 1,4,[5],12:i:-, Infantis, and Schwarzengrund were not detected as frequently in breeder samples, but were found in samples taken during later processing. The authors consider that processes aimed at reduction of Salmonella during processing are effective at reducing Salmonella Kentucky, but less effective at reducing other serovars present in breeder flocks, which were undetected due to limitations in culture selection of only the dominant serovar.
Siceloff and colleagues21 also applied CRISPR-SeroSeq to investigate the prevalence of multiple Salmonella serovars in poultry samples from Georgia between 2020 and 2021 and found that Salmonella Kentucky was the most common out of 134 samples tested, which aligned with the surveillance results. However, the CRISPR-SeroSeq approach also identified Salmonella Cerro and Salmonella Mbandaka as the next most common, as well as serovars Salmonella Enteritidis, Infantis, Montevideo, Thompson, and Typhimurium. In particular, Salmonella Infantis was detected in 12 samples; in 11 of these, it was the minority serovar (second most abundant after dominant serovar). The authors point out that the results suggest that antimicrobial interventions aimed at reduction of Salmonella serovars were effective on the identified dominant serovar. However, the lack of a complete picture of Salmonella serovars present in breeder flocks creates an opportunity for minority serovars to elude antimicrobial mitigation strategies.
Successful application of the CRISPR-SeroSeq method was also demonstrated in a large-scale study conducted in Ontario, Canada, to survey Salmonella serovars on poultry farms across the province.3 The CRISPR-SeroSeq method was used to analyze 442 Salmonella-positive (by PCR) environmental samples collected from 192 poultry farms, and revealed a comprehensive picture of Salmonella serovars on the farms. The samples were taken over a period of ten months from ventilation fans (n = 178), egg belts (n = 96), manure belts (n = 87), floors (n = 37) and other sites (n = 44). A total of 25 serovars were detected in 430 of the samples, with 73.1 percent of the samples containing multiple (up to seven) serovars in a single sample. The most common serovars identified on the farms were Salmonella Kiambu (55.7 percent), Infantis (48.4 percent), Kentucky (27.1 percent), Livingstone (26.6 percent) and Mbandaka/Montevideo (23.4 percent). Different serovars were distributed over ten producer geographical zones across the province (Figure 1).
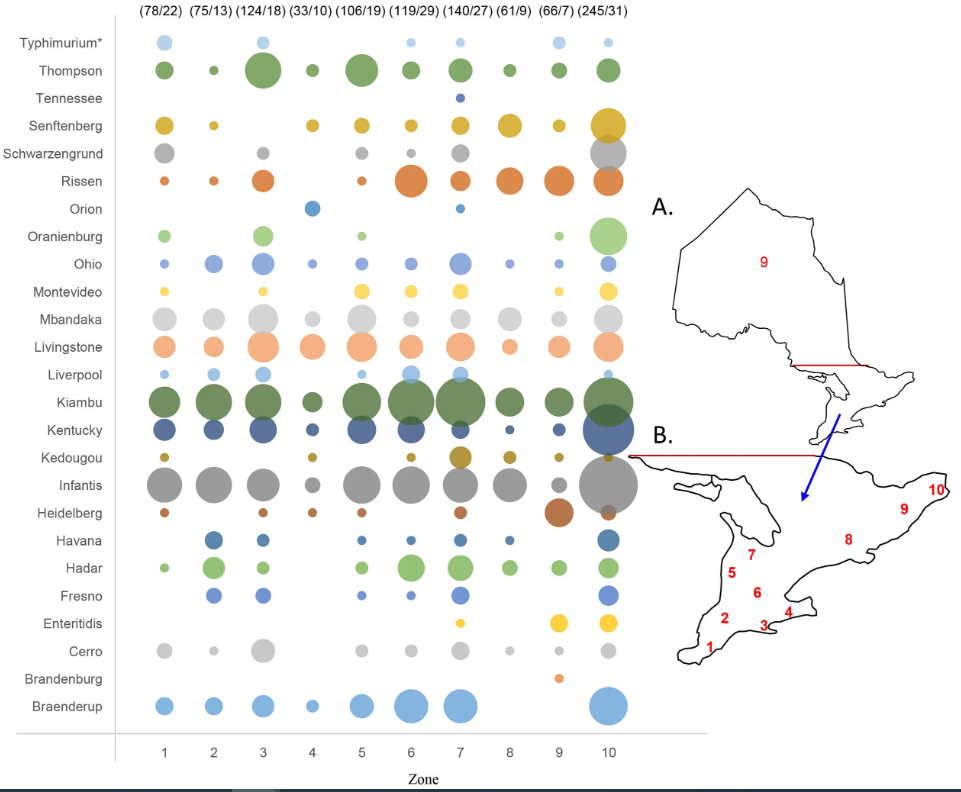
Salmonella Kiambu and Infantis were the most common across the different geographical zones. Salmonella Rissen was detected more often in northern zones as compared with southern zones. Within a farm, different sampling surfaces were found to carry different serovars, although 1–2 common serovars were detected from all surface types sampled based on examination of a limited number of farms. The ventilation fan samples resulted in a higher average number (2.7) of serovars detected per sample as compared with the overall average (2.4). More serovars were detected in warm months (June–October), with an average number of 2.6 serovars detected per sample as compared with cold months (November–March), with an average number of 2.1 serovars detected. Overall, the CRISPR-SeroSeq method detected Salmonella serovars, on average, 3.3 times more frequently than those detected by traditional culture. In 616 instances among 384 samples that were serotyped traditionally, a serovar detected by CRISPR-SeroSeq was missed in the same sample by culture. This included Salmonella Enteritidis, the primary serovar of concern to egg farms, which was detected by CRISPR-SeroSeq and missed by culture among one of the three occasions when it was detected among 5,392 environmental samples tested over a ten-month period.
The most common Salmonella serovars (Kiambu, Infantis, Kentucky, Livingstone, and Mbandaka/Montevideo) detected by CRISPR-SeroSeq in the Ontario study reflect a significant change in predominant serovars, as reported previously. A study of pullet layer and pullet farms from 2002–2003 in Ontario identified serovars Salmonella Heidelberg, Typhimurium, Thompson, Schwarzengrund, and Agona as the most common.22 A subsequent study of egg layer and pullet grower operations in Ontario from 2009–2010 again identified Salmonella Heidelberg (50.7 percent) as the most prevalent serovar, followed by Thompson, Schwarzengrund, Agona, and Kentucky.23 A more recent temporal study of fluff samples from Ontario poultry hatcheries from 2009–2018 revealed Salmonella Kentucky, Enteritidis, Heidelberg, and Senftenberg as the most frequently isolated serovars.24
One of the most significant changes is Salmonella Heidelberg, which showed high prevalence in previous studies but was detected in only 5 percent of farms in the authors' recent study.3 A decrease in Salmonella Heidelberg prevalence in poultry was also observed from 2016 to 2020 in a study conducted in the U.S.21 Another significant change is Salmonella Kiambu, which emerged as the most frequently detected serovar by CRISPR-SeroSeq and the third most frequently detected serovar by culture on Ontario poultry farms,3 although it was under-reported previously. This occurrence is of particular concern since Salmonella Kiambu was implicated in 23.5 percent (50/213) of the 2017 Salmonella outbreaks in the U.S.25 Salmonella Rissen was another emerging serovar in the authors' study, with a detection rate of over 21 percent on the farms by CRISPR-SeroSeq, but it was detected in just 1 percent of farms by culture.3 Significant underestimation of Salmonella Rissen previously or by culture is of concern since Salmonella Rissen has also been recognized as an under-reported and emerging serovar among humans in different countries.26
Toward more Effective Monitoring of Salmonella on Poultry Farms
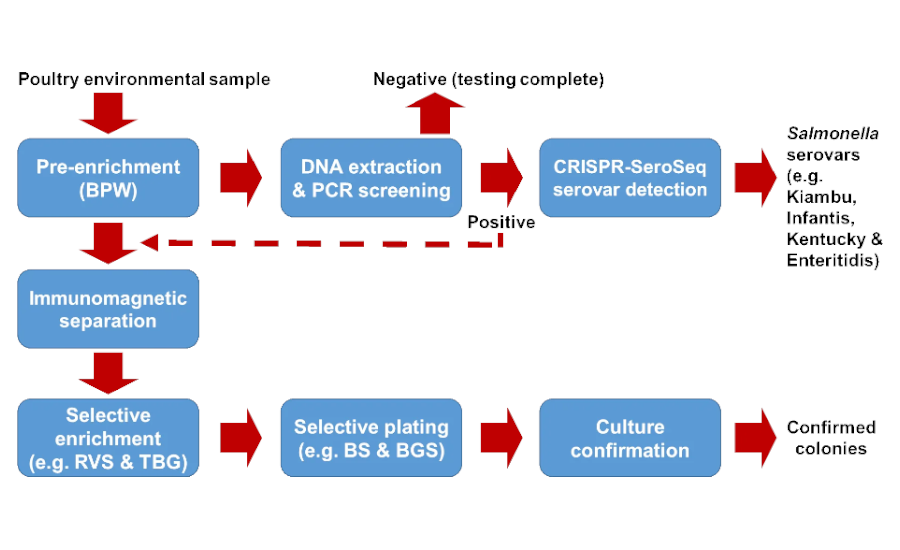
With the availability of numerous PCR methods and the validated CRISPR-SeroSeq method, it is now feasible to detect Salmonella serovars without culture isolation to improve current Salmonella monitoring programs on poultry farms or production facilities. An overall testing approach is depicted in Figure 2. Poultry environmental samples are first pre-enriched in buffered peptone water (BPW) for 18–24 hours. The pre-enrichments are then subjected to real-time PCR to screen for the presence of Salmonella. PCR-positive samples are analyzed via CRISPR-SeroSeq to identify Salmonella serovars contained in the samples. CRISPR-SeroSeq can deliver serovar results within 36–48 hours from DNA extracts of the PCR-positive samples. When culture confirmation is needed (e.g., upon detection of a critical serovar, such as Salmonella Enteritidis, or for outbreak investigations), PCR-positive pre-enrichments can be subjected to immunomagnetic separation (IMS) to concentrate the target cells, using magnetic beads covalently bonded with antibodies specific to Salmonella species, or to a selected subgroup (such as Group D) of Salmonella enterica. The Salmonella cell concentrates are then enriched in a secondary selective medium, such as RVS, TBG, or other selective media, for 18–24 hours. The secondary enrichments can also be subjected to CRISPR-SeroSeq to identify Salmonella serovars, or subjected to culture isolation by plating onto selective agars. Suspect Salmonella colonies are confirmed by biochemical reactions or by PCR. Confirmed colonies are serotyped traditionally or by WGS. In this case, a high number of target colonies from each presumptive Salmonella positive plate must be tested to increase the detection rate if all Salmonella serovars or less abundant serovars need to be confirmed culturally.
Summary
Salmonella outbreaks, poultry diseases, and poultry product contamination events associated with various Salmonella serovars emphasize the need for monitoring all serovars in the poultry production chain. This monitoring will help gain insights into serovar dynamics and emerging serovars, and ensure detection of serovars of critical concern to help improve mitigation practices. Environmental samples from poultry farms often readily reveal the presence of Salmonella in flocks, the ultimate source.
Rapid PCR screening methods are readily available for detection purposes. Detecting all Salmonella serovars routinely via advanced metagenomics tools is technically feasible at present in a limited number of food safety testing laboratories. Increased implementation of the available technologies and tools in monitoring programs—after proposed regulatory framework changes, specific guidelines, and protocols are defined—is expected to help reduce Salmonella infections linked to poultry products.
References
- WHO. "WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden." 2016. Epidemiology Reference Group 2007–2015.
- Diaz, D., P. E. Hernandez-Carreño, D. Z. Velazquez, et al. "Prevalence, main serovars and anti-microbial resistance profiles of non-typhoidal Salmonella in poultry samples from the Americas: A systematic review and meta-analysis." Transboundary and Emerging Diseases 69 (2022): 2544–2558.
- Quinn, M. W., N. F. Linton, C. G. Leon-Velarde, and S. Chen. "Application of a CRISPR Sequence-Based Method for a Large-Scale Assessment of Salmonella Serovars across Ontario Poultry Production Environments." Applied and Environmental Microbiology (February 28, 2023).
- Gantois, I., R. Ducatelle, F. Pasmans, et al. "Mechanisms of egg contamination by Salmonella Enteritidis." FEMS Microbiology Reviews 33 (2009): 718–738.
- Moffatt, C. R. M., J. Musto N. Pingault, et al. "Recovery of Salmonella enterica from Australian Layer and Processing Environments Following Outbreaks Linked to Eggs." Foodborne Pathogens and Disease 14 (2017): 478–482.
- Craven, S. E., N. J. Stern, E. Line, et al. "Determination of the incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in wild birds near broiler chicken houses by sampling intestinal droppings." Avian Diseases 44 (2000): 715–720.
- Hurst, J. L. and W. R. Ward. "Rats and mice and animal feed—A risk too far?" The Veterinary Journal 162, no. 3 (2001): 163–165.
- Wales, A. M., M. Breslin, and R. Davies, R. "Semiquantitative assessment of the distribution of Salmonella in the environment of caged layer flocks." Journal of Applied Microbiology 101 (2006): 309–318.
- Berchieri Jr., A., C. K. Murphy, K. Marston, and P. A. Barrow. "Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: Effect of bacterial and host genetic background." Avian Pathology 30, no. 3 (2001): 221–231.
- Dar, M. A., S. M. Ahmad, S. A. Bhat, et al. "Salmonella Typhimurium in poultry: A review." World's Poultry Science Journal 73 (2017): 345–354.
- Grimont, P. A. and F. X. Weill. "Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella 9 (2007): 1–166.
- Cox, N. A., M. E. Berrang, S. L. House, et al. "Population Analyses Reveal Preenrichment Method and Selective Enrichment Media Affect Salmonella Serovars Detected on Broiler Carcasses." Journal of Food Protection 82, no. 10 (2019): 1688–1696.
- Larsen, B. R., K. E. Richardson, T. Obe, C. Schaeffer, and N. W. Shariat. "Mixed Salmonella cultures reveal competitive advantages between strains during pre‐enrichment and selective enrichment." Journal of Food Safety 41, no. 6 (2001): e12934
- Singer, R. S., A. E. Mayer, T. E. Hanson, and R. E. Isaacson. "Do microbial interactions and cultivation media decrease the accuracy of Salmonella surveillance systems and outbreak investigations?" Journal of Food Protection, 72, no. 4 (2009): 707–713.
- Muniesa, M., A. R. Blanch, F. Lucena, and J. Jofre. "Bacteriophages may bias outcome of bacterial enrichment cultures." Applied and Environmental Microbiology 71, no. 8 (2005): 4269–4275.
- Nadin-Davis, S., L. Pope, D. Ogunremi, B. Brooks, and J. Devenish. "A real-time PCR regimen for testing environmental samples for Salmonella enterica subsp. enterica serovars of concern to the poultry industry, with special focus on Salmonella Enteritidis." Canadian Journal of Microbiology 65, no. 2 (2019): 162–173. https://doi.org/10.1139/cjm-2018-0417
- Ashton, P. M., S. Nair, T. M. Peters, J. A. Bale, D. G. Powell, and A. Painset. "Identification of Salmonella for public health surveillance using whole genome sequencing." PeerJ 4 (2016): e1752.
- Allard, M. W., R. Bell, C. M. Ferreira, et al. "Genomics of foodborne pathogens for microbial food safety." Current Opinion in Biotechnology 49 (2018): 224–229.
- Vincent, C., V. Usongo, C. Berry, et al. "Comparison of advanced whole genome sequence-based methods to distinguish strains of Salmonella enterica serovar Heidelberg involved in foodborne outbreaks in Quebec." Food Microbiology 73 (2018): 99–110.
- Thompson, C. P., A. N. Doak, N. Amirani, et al. "High-Resolution Identification of Multiple Salmonella Serovars in a Single Sample by Using CRISPR-SeroSeq. Applied and Environmental Microbiology 84, no. 21 (2018): e01859-18.
- Siceloff, A. T., D. Waltman, and N. W. Shariat. "Regional Salmonella Differences in United States Broiler Production from 2016 to 2020 and the Contribution of Multiserovar Populations to Salmonella Surveillance." Applied and Environmental Microbiology 88, no. 8 (2022): e0020422.
- Leon-Velarde, C. G., H. Y. Cai, C. Larkin, et al. "Evaluation of methods for the identification of Salmonella enterica serotype Typhimurium DT104 from poultry environmental samples." Journal of Microbiological Methods, 58, no. 1 (2004): 79–86.
- Leon-Velarde, C. G., S. Chen, F. Olea-Popelka, and J. Odumeru. "Characterization and distribution of Salmonella from egg layer and pullet grower operations in Ontario, Canada, by MLVA." The 91st Conference of Research Workers in Animal Diseases. December 5–7, 2010, Chicago, Illinois, U.S.
- Murray, C. E., C. Varga, R. Ouckama, and M. T. Guerin. "Temporal Study of Salmonella enterica Serovars Isolated from Fluff Samples from Ontario Poultry Hatcheries between 2009 and 2018." Pathogens, 11, no. 1 (2021): 9.
- Hassan, R., B. Whitney, D. L. Williams, Outbreak Investigation Team, et al. "Multistate outbreaks of Salmonella infections linked to imported Maradol papayas—United States, December 2016–September 2017." Epidemiology and Infection 147 (2019): e265.
- Elbediwi, M., D. Shi, S. Biswas, X. Xu, and M. Yue. "Changing Patterns of Salmonella enterica Serovar Rissen From Humans, Food Animals, and Animal-Derived Foods in China, 1995–2019." Frontiers in Microbiology 12 (2021): 702909.




.webp?height=200&t=1691503719&width=200)


