Faster, Cheaper Sequencing Transforms Microbial Food Safety Testing
Transformational insights in microbial surveillance technology

New advances in sequencing technology could help transform the way microbial food safety is managed both within our own laboratory network and throughout the global supply chain. The implementation of rapid whole-genome sequencing (WGS) could help transform microbial risk surveillance across the food industry from a factory and ingredient surveillance approach to a more preventive approach—one in which we can identify outbreak indicators to predict, and take steps to prevent, a problem before it even occurs. The reduction in cost per sample, while maintaining a practical testing time, could provide vital tools during incident investigations, enabling manufacturers to make rapid, informed decisions on product releases to ensure the supply chain—and therefore the consumer—is safe from microbial contamination.
Why Research Whole-Genome Sequencing?
Food is more likely to be contaminated with microbes along the farm-to-table route. The U.S. Centers for Disease Control and Prevention estimates that approximately 35 million infections, 26,500 hospitalizations, and 420 deaths are caused each year in the Unites States by food contaminated with the microbe Salmonella.1 The scale of the impact of these contamination events, not just on the public but also on businesses through costly recall campaigns, highlights the need to reinforce control measures in the food industry, including faster and more accurate confirmation and tracking methods.
Since the 1960s, researchers have distinguished between different strains of Salmonella through a classification system known as serotyping. Serotypes are groups within a single species of microorganisms, such as bacteria or viruses, that share distinctive surface structures.
Conventional methods of serotyping Salmonella are phenotypic, meaning they are related to observable characteristics. There are two main classification criteria related to the serotype naming process in Salmonella: the O antigen, which can be found on the surface of the bacteria, and the H antigen, which can be found on the flagellum (the tail). The combination of the information of these two pieces, together with the surface antigen Vi that can be found in some species of Salmonella, contributes the bulk of what is known as the “antigenic formula.”
Visually distinguishing between more than 100 different types of flagella is a time-consuming process, requiring well-trained personnel2 to conduct and obtain reliable results. All told, there are over 2,600 known serotypes of Salmonella,3 and as such, a more granular approach was thought to be required.
Recent Innovations in Whole-Genome Sequencing
Looking for quick answers on food safety topics?
Try Ask FSM, our new smart AI search tool.
Ask FSM →
While several molecular methods,4 such as microarray-based subtyping,5 have been developed to overcome the drawbacks of traditional serotyping methods, it is serotyping using WGS data that is being increasingly used by regulators, industry, and academia to identify the source of outbreaks, investigate their contamination pathway, conduct routine surveillance, and research further purposes.
In WGS, serotypes are differentiated based upon their genetic code; researchers identify fragments of genetic code that can be used as markers specific to a particular Salmonella serotype or variant. At the Mars Global Food Safety Center (GFSC), we have been collaborating with experts at the Center for Food Safety, University of Georgia, and the Department of Food Science, Cornell University, to evaluate and develop methodologies using state-of-the-art WGS nanopore platforms that can analyze any length of DNA in real time, providing the potential for more rapid microbiological analysis than ever before. This electronics-based technology is currently being used in more than 100 countries worldwide, and we are now formally testing it in the field of food safety.
Advances in Sequencing
Based on the latest research, we believe that a significant reduction in the time taken to confirm pathogen contamination and track the pathogen pathway could provide vital information during incident investigations, enabling manufacturers to make informed decisions on product releases to ensure the supply chain is safe from contamination. A new advance in sequencing technology to facilitate this could help transform the way microbial food safety is managed within both global laboratory networks and the raw material food supply chain.
Over the past 3 years, we have worked with our academic partners to optimize the sequencing technology for routine surveillance—focusing on ensuring accurate and timely in-house Salmonella serotyping. We have previously demonstrated how we can reduce the turnaround time per sample from 1 to 2 weeks down to less than 1 day.6
The Power of DNA Barcoding
We have been investigating the practicality of using nanopore WGS to simultaneously sequence multiple isolates of Salmonella in one flow cell—known as multiplex sequencing.7 When running a WGS test, sequencing reads must be recombined into longer stretches of code. The process of determining the correct order of multiple sequenced DNA reads to reconstruct the original DNA sequence of the genome as close as possible is known as genome assembly.
Genome assembly of WGS data is further complicated when pooling multiple samples together—it can be imagined as similar to attempting to complete five 100-piece puzzles that have all been placed within the same 500-piece box. Even with images to compare the pieces against, the task is nontrivial compared with constructing only one 100-piece puzzle on its own. To help simplify the task, the puzzle manufacturer could place a marker (e.g., a colored dot) on the back of some of the puzzle pieces prior to its being broken up—the fragments could then be categorized according to their markers, and the puzzle assembled around each of the known pieces. In WGS analysis, this process is called DNA barcoding.
This is how DNA barcoding facilitates high-throughput sequencing when working with large quantities of complexed samples via multiplex sequencing.
How Multiplex Sequencing Works
In multiplex sequencing (Figure 1), the bacterial samples are grown into colonies to ensure there is sufficient DNA for testing (Figure 1, step 1) before the DNA is extracted and a set of oligonucleotide tags of known DNA sequences (barcodes) are added to the DNA fragments during sequencing (Figure 1, step 2). A set number of samples are then pooled, with mixing to ensure homogeneity, and library preparation protocols are simultaneously prepared so that each read can later be identified and sorted before genome assembly (Figure 1, step 3). Aliquots are taken from the pooled batch (Figure 1, step 4) and run on the flow cells in the sequencing platform (Figure 1, step 5). Once sequenced, advanced computational algorithms are used to “demultiplex” the DNA. The demultiplexed sequencing data then go through genome assembly and analysis of in silico serotype prediction by comparing the assembled genomes with a database of serotype-determinant loci or other genomic markers or patterns for serotype prediction (Figure 1, step 6).
FIGURE 1. Multiplex Nanopore Sequencing in Six Steps
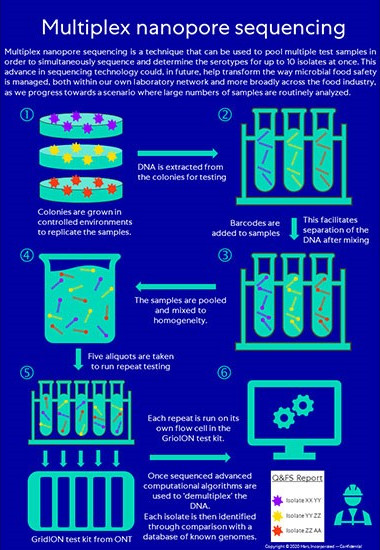
Validation for the Food Industry
To ensure the protocol met the needs of the food industry for accurate, rapid, and cost-efficient Salmonella confirmation and serotype classification, the method was evaluated as to the serotype-prediction accuracy by using WGS data from multiplex nanopore sequencing with 3, 4, 5, 7, or 10 Salmonella isolates, with each isolate representing one Salmonella serotype, pooled in a single flow cell.
Ten strains of bacteria, including common strains such as Enteritidis and Senftenberg, as well as strains known from the literature to be difficult to predict [Paratyphi B (dt+)] and difficult to differentiate [Typhimurium vs. Typhimurium (O5–)], were chosen to ensure the proposed methodology is robust for all likely situations encountered. Two in silico serotype prediction tools were used, with both accurately predicting all the multiplexed isolates under each multiplexing strategy (when depth of genome coverage ≥ 50× for each isolate)—demonstrating in principle that multiplexing of Salmonella using nanopore WGS can be achieved.
Cost Savings at Scale
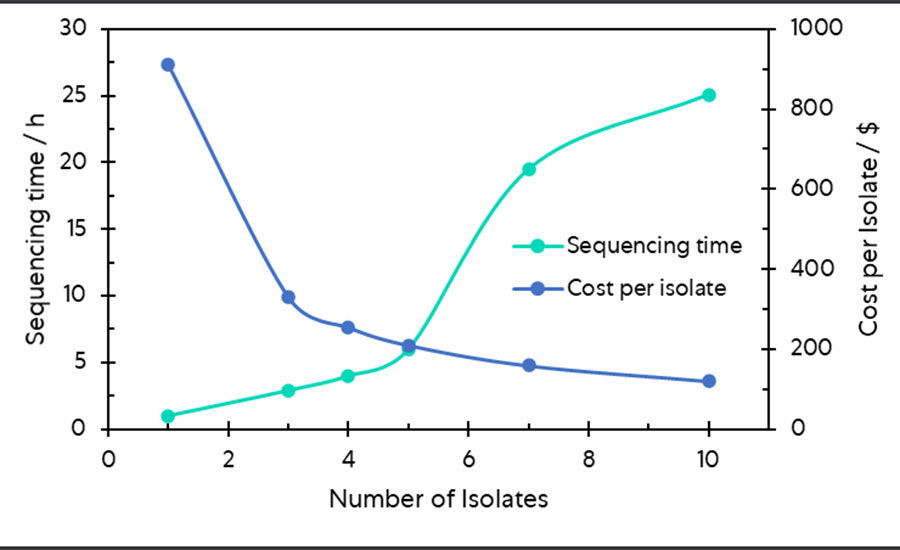
The time requirement and cost per sample of completing a sample run are typically the driving force behind WGS research for routine use. While more research is required, this pioneering study successfully demonstrated that multiplex sequencing employing nanopore WGS could be used to simultaneously predict serotypes for between 3 and 10 Salmonella isolates, with the protocol optimized at 5 isolates within an average of more than 6 hours of sequencing (Figure 2). The newly proposed methodology constrains the time requirements to run the test to within 1 working day to suit laboratory worker requirements, while reducing the cost per isolate to 23 percent of that incurred with single-nanopore sequencing, paving the way for routine and batch testing within factory networks.
FIGURE 2. The Payoff between Cost and Time Savings
Transforming the Future of Food Safety
At the Mars GFSC, we believe we have a clear responsibility to lead in food safety. However, we also believe that no one entity can do this alone. That is why we are taking a truly collaborative approach to food safety, one rooted in fostering mutual sharing and collaboration. Part of that commitment involves driving research and collaboration to move toward faster detection, identification, and, ultimately, prediction of microbial contamination risks.
Using multiplex WGS data generated by a long-read sequencing platform provides the food industry, food authorities, and regulators a possible alternative to short-read sequencing platforms for serotype prediction for pathogen surveillance. It will also guide the feasibility of applying this technology in the food and public health arenas to improve Salmonella confirmation, subtyping, and source tracking efficiency by shortening the turnaround time and increasing the identification accuracy compared with conventional serotyping methods, with improved cost-efficiency.
By demonstrating that multiplex sequencing of five isolates can dramatically reduce the cost per isolate while providing high-quality coverage of the genome and high-accuracy prediction within a single working day, we are one step closer to routine implementation of this technique during incident investigations. The use of WGS throughout the global supply chain will significantly improve the efficiency of microbial contamination source tracking and bring us nearer our goal of safe food for all.
Read more from the Mars Global Food Safety Center at our website, gfsc.mars.com, or follow us on LinkedIn at linkedin.com/showcase/marsgfsc/.
References
- https://www.cdc.gov/salmonella/.
- https://www.tandfonline.com/doi/abs/10.3109/1040841X.2013.837862.
- https://www.sciencedirect.com/science/article/abs/pii/S0923250814001065.
- https://www.frontiersin.org/articles/10.3389/fmicb.2019.02554/full.
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3043481/.
- https://www.sciencedirect.com/science/article/pii/S0740002020300411.
- https://www.frontiersin.org/articles/10.3389/fmicb.2021.637771/full.
Recent Publications from the Mars Global Food Safety Center on WGS
- Wu, X., et al. 2021. “Evaluation of Salmonella Serotype Prediction With Multiplex Nanopore Sequencing.” Front Microbiol 12: 380–393.
- Xu, F., et al. 2021. “Evaluation of Nanopore Sequencing Technology to Identify Salmonella enterica Choleraesuis var. Kunzendorf and Orion var. 15+, 34+.” Int J Food Microbiol 346: 109167.
- Xu, F., et al. 2020. “Evaluation of Real-Time Nanopore Sequencing for Salmonella Serotype Prediction.” Food Microbiol 89: 103452.
Silin Tang, Ph.D., is a senior research scientist at the Mars Global Food Safety Center, where she leads research projects on the development, evaluation, and validation of pathogen detection and identification methods for the food industry, including the application of WGS technologies in source tracking of foodborne pathogens. She graduated from Cornell University with a Ph.D. in food science and technology and has more than 10 years of research experience working in microbiological food safety.
.webp?height=96&t=1634598143&width=96)







